Last update: 20250102
Design release DNA SNV INDEL v1
Protocol name: design_dna_snv_indel_v1_relsease (this document) for design_dna_snvindel_v1 (see design_dna_snvindel_v1).
Release information
Current release version: v1
Location temp:/project/data/shared_all/release_dna_snv_indel_v1
Location stable:/project/data/shared/release_dna_snv_indel_v1
Status
Status:
design_dna_snv_indel_v2_relseaseis currently recommended.
We have added new annotation settings affecting which QV are reported.
Any modifications due to error will be notified and a new release ID will be issued (e.g.design_dna_snv_indel_v1.2_relsease).
About
If you are reading this then you are interested in using the release data from the design_dna_snvindel_v1 pipeline. This pipeline produces a multi-use dataset via the qualifying variants 1 (QV1) protocol. Incremental additions with new data for this release will be added. Potential uses are suggested as the end-points in figure 1 (e.g. statistical genomics, etc.)
The protocol used was:
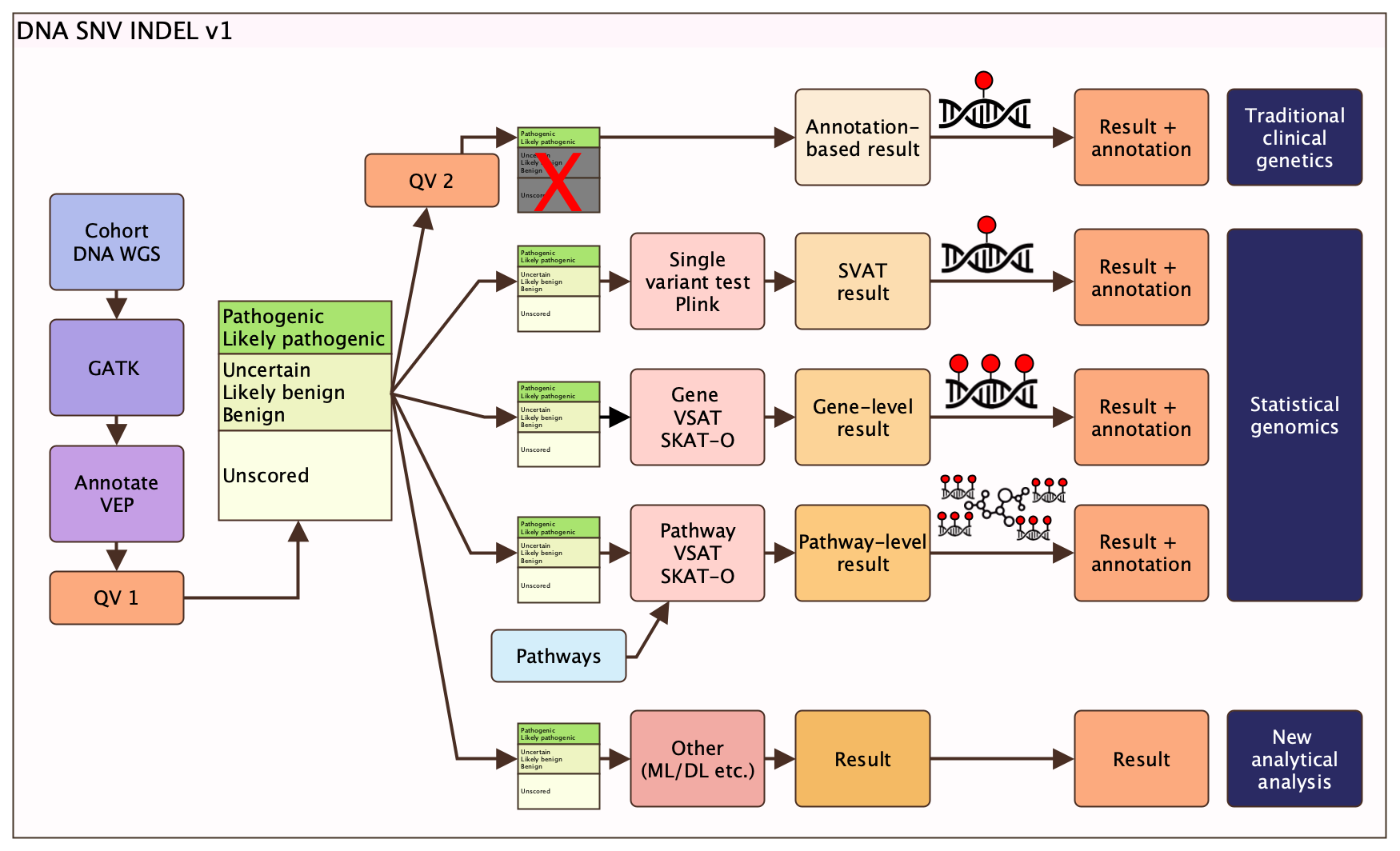
Figure 1: Summary of design DNA SNV INDEL v1 pipeline plan.
Contents
See the README.md for this release to find files:
dna_snv_indel_v1_release.README.md
Data synchronisation process
- Sync release data to
shared_all. - Log and verify data during sync to
shared_all. - Data manager syncs from
shared_alltoshared. - The final release is locked in
sharedbut allows incremental additions. - Labelled as
release_dna_snv_indel_v1.
Data Availability
All raw and processed data are accessible to all internal researchers. Processed/intermediate data might change during projects, so released data is recommended for downstream projects. Release data should have detailed supporting documentation following the protocol. Essential data is curated to minimise storage use. Non-release data is accessible as usual from user directories but may be overwritten or updated without notice. Data paths are listed in the release script, generally read from a master list of variables.
Current version script
release_dna_snv_indel_v1.sh is run from the user src directory.
Checksums
All files are logged with their md5 checksum. To verify integrity run: md5sum -c checksums.md5
$ cat checksums.md5
457dc93749669bed628575dfe551bdac ./release_dna_snv_indel_v1.sh.log
d35670b5ac26302b19f07c4cf3a96977 ./release_dna_snv_indel_v1.out
$ md5sum -c checksums.md5
./release_dna_snv_indel_v1.sh.log: OK
./release_dna_snv_indel_v1.out: OK
Stats
Chromosome 1
Number of samples: 180
Number of SNPs: 1543167
Number of INDELs: 385213
Number of MNPs: 0
Number of others: 0
Number of sites: 1972696
Chromosome 2
Number of samples: 180
Number of SNPs: 1620394
Number of INDELs: 394968
Number of MNPs: 0
Number of others: 0
Number of sites: 2061648
Chromosome 3
Number of samples: 180
Number of SNPs: 1316780
Number of INDELs: 324975
Number of MNPs: 0
Number of others: 0
Number of sites: 1673128
Chromosome 4
Number of samples: 180
Number of SNPs: 1337064
Number of INDELs: 316959
Number of MNPs: 0
Number of others: 0
Number of sites: 1689336
Chromosome 5
Number of samples: 180
Number of SNPs: 1196261
Number of INDELs: 293793
Number of MNPs: 0
Number of others: 0
Number of sites: 1520849
Chromosome 6
Number of samples: 180
Number of SNPs: 1214390
Number of INDELs: 303459
Number of MNPs: 0
Number of others: 0
Number of sites: 1552036
Chromosome 7
Number of samples: 180
Number of SNPs: 1145244
Number of INDELs: 277881
Number of MNPs: 0
Number of others: 0
Number of sites: 1460406
Chromosome 8
Number of samples: 180
Number of SNPs: 1020218
Number of INDELs: 240781
Number of MNPs: 0
Number of others: 0
Number of sites: 1284467
Chromosome 9
Number of samples: 180
Number of SNPs: 858094
Number of INDELs: 198839
Number of MNPs: 0
Number of others: 0
Number of sites: 1080126
Chromosome 10
Number of samples: 180
Number of SNPs: 946539
Number of INDELs: 237079
Number of MNPs: 0
Number of others: 0
Number of sites: 1212448
Chromosome 11
Number of samples: 180
Number of SNPs: 910215
Number of INDELs: 221252
Number of MNPs: 0
Number of others: 0
Number of sites: 1154276
Chromosome 12
Number of samples: 180
Number of SNPs: 903797
Number of INDELs: 242211
Number of MNPs: 0
Number of others: 0
Number of sites: 1173553
Chromosome 13
Number of samples: 180
Number of SNPs: 683880
Number of INDELs: 169236
Number of MNPs: 0
Number of others: 0
Number of sites: 872669
Chromosome 14
Number of samples: 180
Number of SNPs: 618962
Number of INDELs: 156463
Number of MNPs: 0
Number of others: 0
Number of sites: 792576
Chromosome 15
Number of samples: 180
Number of SNPs: 609207
Number of INDELs: 141670
Number of MNPs: 0
Number of others: 0
Number of sites: 768672
Chromosome 16
Number of samples: 180
Number of SNPs: 653752
Number of INDELs: 151086
Number of MNPs: 0
Number of others: 0
Number of sites: 826293
Chromosome 17
Number of samples: 180
Number of SNPs: 521864
Number of INDELs: 161194
Number of MNPs: 0
Number of others: 0
Number of sites: 700410
Chromosome 18
Number of samples: 180
Number of SNPs: 534571
Number of INDELs: 130405
Number of MNPs: 0
Number of others: 0
Number of sites: 680157
Chromosome 19
Number of samples: 180
Number of SNPs: 455618
Number of INDELs: 136628
Number of MNPs: 0
Number of others: 0
Number of sites: 610238
Chromosome 20
Number of samples: 180
Number of SNPs: 427477
Number of INDELs: 108222
Number of MNPs: 0
Number of others: 0
Number of sites: 547588
Chromosome 21
Number of samples: 180
Number of SNPs: 333850
Number of INDELs: 67464
Number of MNPs: 0
Number of others: 0
Number of sites: 413496
Chromosome 22
Number of samples: 180
Number of SNPs: 332138
Number of INDELs: 74089
Number of MNPs: 0
Number of others: 0
Number of sites: 418913
Chromosome X
Number of samples: 180
Number of SNPs: 678392
Number of INDELs: 183618
Number of MNPs: 0
Number of others: 0
Number of sites: 881808
Chromosome Y
Number of samples: 180
Number of SNPs: 71640
Number of INDELs: 1289
Number of MNPs: 0
Number of others: 0
Number of sites: 77666