Last update: 20241216
DNA annotation
Variant annotation is a critical step in clinical and statistical genetics. Popular tools for applying annotation data to VCF format genetic data include:
- Variant Effect Predictor (VEP) link: VEP
- NIRVANA link: NIRVANA
- ANNOVAR link: ANNOVAR
We are using VEP with Conda, but we are likely to test additional methods (licence). Additionally, these tools must be paired with a set of data sources containing the annotation information which will be applied to each variant.
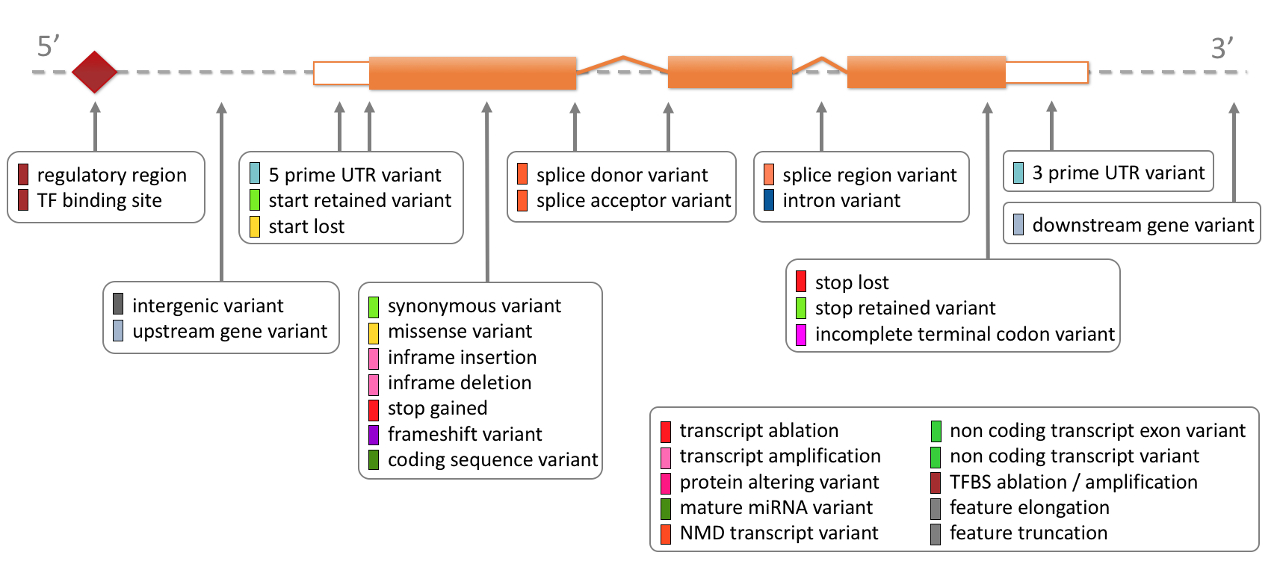
The variant consequence may be one of the defining criteria by which variants can be included in analysis since they are interpretable or of ostensibly known significance.
The consequences provided by VEP can provide a simple reference example to understand its function. For example, HIGH impact variants might be a likely consequence for identifying candidates disease-causing: Ensembl Variation - Calculated variant consequences.\
You may have observed cases in literature where clinical research reporting relied on variant effect consequence alone for known disease-related genes, but this practice is likely to introduce spurious results. It is important to use established criteria for selecting consequences of interest combined with additional filtering methods to define evidence thresholds. See the ACMG interpretation standards for examples.